Variation between long-diverged fungal species.
Fungi specialize to their environment in amazing ways. When a fungal species from one niche differs radically from its relatives elsewhere, we would love to know the mechanism at play. In many cases the underlying adaptive alleles are a mystery, especially if they arose long ago. The main roadblock to date has been that most experimental tools in evolutionary genetics research are only applicable to comparison across strains, not species. Ongoing work in the lab is devoted to breaking through this limitation. We developed a novel approach to map the genetic basis of differences between distantly related species through reciprocal hemizygote analysis of interspecific hybrids. 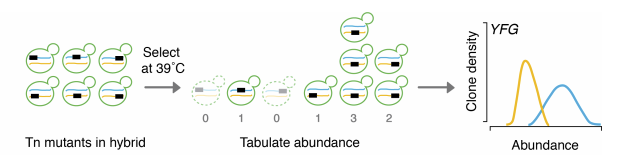
A transposon (rectangle) disrupts the allele from S. cerevisiae (blue) or S. paradoxus (orange) of a gene (YFG) in an interspecific hybrid (green). Clones lacking the prothermotolerance allele grow poorly at 39°C (dashed outlines), as measured by sequencing and reported in smoothed histograms (traces, colored to indicate the species’ allele that is not disrupted)
Our method reveals the nucleotide-level variants that a given fungus has used to adapt to its environment over evolutionary time. The pilot project uses thermotolerance in Saccharomyces yeast as a testbed, with the promise of extension to various phenotypes in many other eukaryotes. For more information, see our recent publication on the new technique, RH-Seq, or the the video below:
Mapping genotype to phenotype in fungi.
Most fungal genomes are poorly annotated, so we have no idea of the genetic architecture for many fungal traits of industrial and biomedical relevance. In published work, our lab has used variation across wild populations to find the molecular basis of morphology traits in the model fungi Saccharomyces and Neurospora; we have also developed genomic screening approaches to annotate candidate virulence islands in the plant pathogen Zymoseptoria and lipid metabolism genes in the oleaginous yeast Rhodosporidium. Our next project, now in progress, is focused on strain variation in virulence and stress resistance traits in the human pathogen Coccidioides.
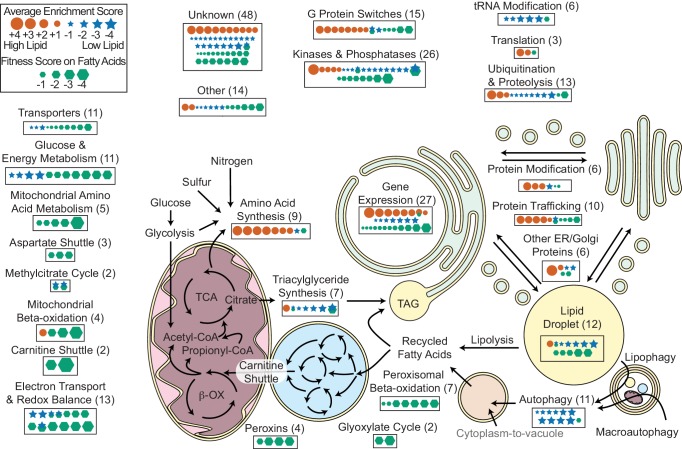
Key metabolic pathways and cellular functions mediating lipid metabolism as identified from fitness scores on fatty acid and enrichment scores from lipid accumulation screens. From Figure 6 of Coradetti et al., 2018
Uncovering the genetics of cellular senescence.
Cellular senescence is an tumor-suppressive stress response characterized by physiologically irreversible growth arrest, apoptosis resistance, and increased secretion of soluble inflammatory and tissue remodeling factors. Senescent cells have been implicated in the development of many aging-related pathologies, and now classic work has identified several major regulators of this response such as p16, p21, NF-kB, and pRB. However, given the complexity and variability of the senescent phenotype, we believe that there are still many molecular mechanisms of cellular senescence that have yet to be discovered. To this end we turn to natural variation in senescence phenotypes between two species of mice; the commonly used house mouse Mus musculus, and its distant relative, the Algerian mouse M. spretus. Our aim is to uncover the genetics behind natural variation in senescent markers between M. musculus and M. spretus via RH-seq and to identify novel regulators of gene expression during senescence through our novel natural variation based transcription factor screening technique.
Genetics of axon damage response and axon regeneration.
The function of the nervous system relies on the transmission of information throughout an organism via projections of neuronal cells. A subset of these projections called axons relay information from one cell to another downstream cell, sometimes over long biological distances. Axons can be severed by traumatic injury; can degrade as part of the etiology of stroke or neurodegenerative diseases; and can be damaged by chemotherapeutic drugs during cancer treatment. Loss of axons also occurs during normal aging, in the absence of disease altogether. Damage to axons can result in aberrant signaling, or a complete loss of signaling if the axon is severed and fails to reconnect. Axon regeneration in the central nervous system (CNS) of mammals is extremely limited, therefore damage to the CNS often results in permanent dysfunction. Currently, there are no approved therapies to regrow damaged CNS axons in humans.
Interestingly, a previous study has demonstrated the remarkable ability of a Southeast Asian subspecies of mouse (Mus musculus castaneus) to regrow CNS axons after injury compared to other mice. We are using this natural variation in axon regeneration phenotypes between the M. m. castaneous and the more commonly used M. m. musculus to study the biology of axon damage response and regeneration. From one angle, we are using RH-seq to screen for the natural genetic variants that underlie the regenerative phenotype of M. m. castaneus. Separately, we are using the natural variation in cis-acting genetic regulatory elements between subspecies to investigate differences in evolutionary selective pressures, and to look for novel gene regulatory networks involved in the cellular response to axon damage.